BtGal95A (exo-α-1,2-L-Galactosidase)
BtGal95A
Ex-Gal0025
(EC.3.2.1) exo-α-1,2-L-Galactosidase
CAZy Family: GH95
PROPERTIES
1.ELECTROPHORETIC PURITY
-Single band on SDS-gel electrophoresis (MW ~93kDa)
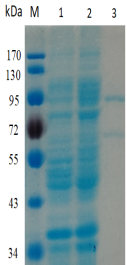
Figure 1. Electrophoresis analysis of BtGal95A. M, molecular weight marker (PageRuler Prestained Protein Ladder, Thermo Scientific); lane 1, culture lysate before IPTG induction; lane 2, culture lysate after IPTG induction; lane 3, BtGal95A purified from Ni sepharose fastflow column.
2.SPECIFIC ACTIVITY
0.95 U/mg protein (on pNP-α-gal) at pH 7.0 and 37°C.
One Unit of pNP-α-gal activity is defined as the amount of enzyme required to release 1 μmol of p-nitrophenyl per minute from pNP-α-gal (5 mM) in phosphate buffer (10 mM) pH 7.0.
3.RELATIVE RATES OF HYDROLYSIS OF SUBSTRATES
Table 1. Relative activity ofBtGal95Aon different substratesa.
Substrateb |
Relative activity (%)c |
pNPαGlc |
_ |
pNPβGlc |
_ |
pNPαGal |
100±0.0 |
pNPβGal |
_ |
pNPαMan |
_ |
pNPβMan |
_ |
pNPαXyl |
_ |
pNPβXyl |
_ |
pNPαAraf |
_ |
pNPαArap |
_ |
pNPαRha |
_ |
pNPαFuc |
_ |
aReactions were performed with 5 mM substrate, pH7.5, at 37°C for 5 min.
bAbsorption caused by released p-nitrophenol was measured at 405 nm. The relative activity on pNPαGal was taken as 100%.
cThe data are reported as means±standard errors from the mean for three independent experiments.
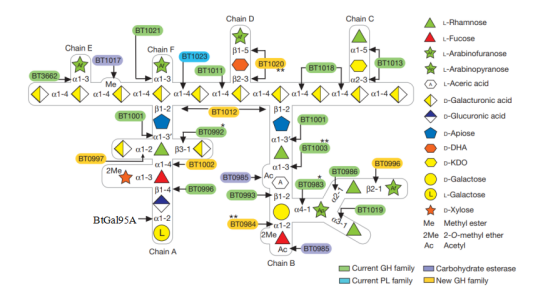
Figure 2. Schematic of enzymes and PULs involved in RG-II degradation. Sugars shown using the Consortium for Functional Glycomics notation25. Enzymes are appropriately colour-coded. Single asterisks denote a new activity for a glycoside hydrolase (GH) family; double asterisks denote enzymes with activities that have not previously been observed. d-KDO, 3-deoxy-d-manno-octulosonic acid; D-DHA, 2-keto-3-deoxy-d-lyxo-heptulosaric acid; PL, polysaccharide lyase.
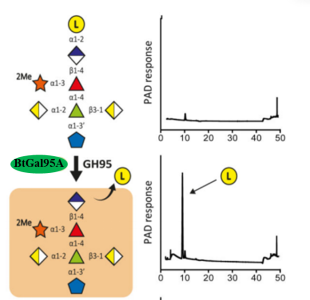
Figure3.BtGal95A-mediated depolymerisation of RG-II Chain A. The substrates comprised Chain A released from RG-II by trifluoroacetic acid.Individual proteins (1 μM) were incubated with the glycans (5 mM) for 16 h at 37 °C in 20 mM sodium phosphate buffer, pH 7.0. Monosaccharides and oligosaccharides generated were identified by HPAEC-PAD and ESI-MS, respectively. Verification of the model was achieved by reconstituting the pathway using the six enzymes in concert, which showed that the GHs only functioned in the order shown in the figure. The example is from technical replicates n = 3.
4.PHYSICOCHEMICAL PROPERTIES
pH Optima:7.5
Temperature Optima:37°C
5.STORAGE CONDITIONS
The enzyme should be stored at -20°C. For assay, this enzyme should be diluted in phosphate buffer (10 mM) pH7.0. Swirl to mix the enzyme immediately prior to use.
6. REFERENCES
[1]Ndeh D, Rogowski A, Cartmell A, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions[J].Nature. 2017,544(7648): 65-70.


