ZgGlc16B(endo-β-1,3-Glucanase)
ZgGlc16B
Ex-Glu0313
(EC.3.2.1.39) endo-β-1,3-Glucanase
CAZy Family: GH16
PROPERTIES
1.ELECTROPHORETIC PURITY
-Single band on SDS-gel electrophoresis (MW ~101kDa)
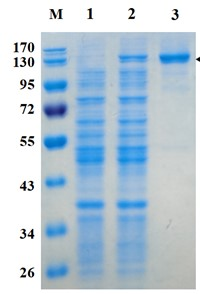
Figure 1. Electrophoresis analysis of ZgGlc16B. M, molecular weight marker (PageRuler Prestained Protein Ladder, Thermo Scientific); lane 1, culture lysate before IPTG induction; lane 2, culture lysate after IPTG induction; lane 3, ZgGlc16B purified from Ni sepharose fastflow column.
2.SPECIFIC ACTIVITY
34.8 U/mg protein (onCurdlan) at pH7.0 and40°C
One Unit of β-glucan activity is defined as the amount of enzyme required to release 1 μmol of glucose from Curdlan (10mg/mL) inphosphate buffer (200 mM) pH7.0.
3.RELATIVE RATES OF HYDROLYSIS OF SUBSTRATES
Table 1. Relative activity of ZgGlc16Bon different substratesa.
Substrate |
Linkage |
ZgGlc16B |
|
Specific activit(U/mg) |
Relative activity(%) |
||
curdlan |
β-1,3 |
34.8 |
100 |
ganoderan |
β-1,3 |
22.1 |
64 |
barley polysaccharide |
β-1,3/1,4 |
15.9 |
46 |
Cellulose polysaccharide |
β-1,4 |
0 |
0 |
Shier polysaccharide |
β-1,6 |
0 |
0 |
starch polysaccharide |
α-1,4 |
0 |
0 |
aReactions were performed with 10 mg/ml (polysaccharides) substrate, pH 7 .0, at 4 0 °C for 5 min.
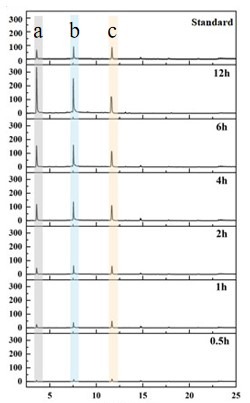
Fig.2. HPAEC detection results of ZgGlc16B action site study. The experiment used gel polysaccharide as substrate. a, monosaccharide. b, disaccharide. c, trisaccharide.
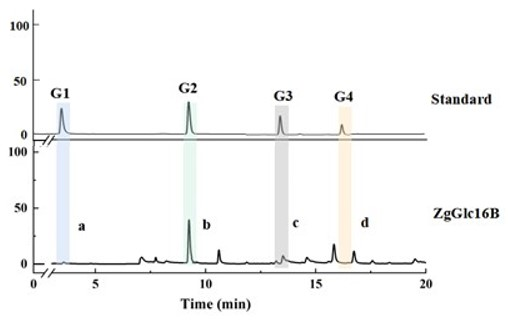
Fig.3. HPAEC diagram of barley polysaccharide hydrolyzed by ZgGlc16B.G1, monosaccharide.G2, Linear glucobiose.G3, Linear glucotriose.G4, Linear glucotetraose.
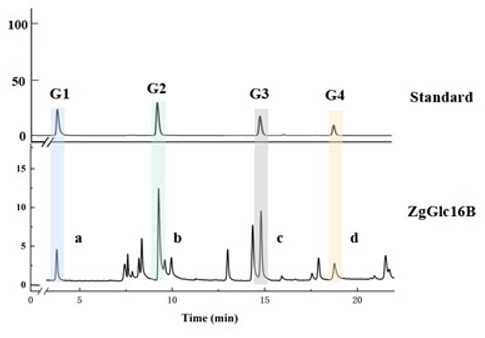
Fig.4. HPAEC diagram of ZgGlc16B after hydrolysis of Ganoderma lucidum polysaccharides. G1, monosaccharide.G2, Linear glucobiose.G3, Linear glucotriose.G4, Linear glucotetraose.
4.PHYSICOCHEMICAL PROPERTIES
pH Optima:6.0
pH Stability: 6.0-9.0
Temperature Optima:40°C
Temperature Stability:<35°C
5.STORAGE CONDITIONS
The enzyme should be stored at -20°C. For assay, this enzyme should be diluted in phosphate buffer (20 mM) pH 7.0. Swirl to mix the enzyme immediately prior to use.


