BtGal35A(exo-β-1,4-Galactosidase)
BtGal35A
Ex-Gal0063
(EC.3.2.1.23)exo-β-1,4-Galactosidase
CAZy Family: GH35
PROPERTIES
1.ELECTROPHORETIC PURITY
-Single band on SDS-gel electrophoresis (MW ~79 kDa)
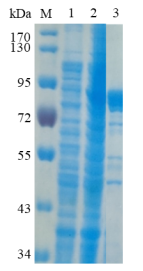
Figure 1. Electrophoresis analysis of BtGal35A. M, molecular weight marker (PageRuler Prestained Protein Ladder, Thermo Scientific); lane 1, culture lysate before IPTG induction; lane 2, culture lysate after IPTG induction; lane 3, BtGal35A purified from Ni sepharose fastflow column.
2.SPECIFIC ACTIVITY
0.29 U/mg protein (on pNP-β-Gal) at pH 7.0 and 37°C.
One Unit of pNP-β-Gal activity is defined as the amount of enzyme required to release 1 μmol of p-nitrophenyl per minute from pNP-β-Gal (5 mM) in phosphate buffer (20 mM) pH 7.0.
3.RELATIVE RATES OF HYDROLYSIS OF SUBSTRATES
Table 1. Relative activity of BtGal35A on different substratesa.
Substrateb |
Relative activity (%)c |
pNPβGlu |
_ |
pNPβGal |
100.0±0.0 |
pNPβMan |
_ |
pNPβXyl |
_ |
pNPαGlc |
_ |
pNPαGal |
70.1±0.7 |
pNPαMan |
_ |
pNPαAraf |
_ |
pNPαArap |
_ |
Sophorose |
_ |
Laminaribose |
_ |
Cellobiose |
_ |
Gentiobiose |
_ |
aReactions were performed with 5mM substrate, pH 7.0, at 37°C for 10 min.
bAbsorption caused by released p-nitrophenol was measured at 405 nm. The relative activity on pNP-β-Gal was taken as 100%.
cThe data are reported as means±standard errors from the mean for three independent experiments.
Table 2.Kinetics ofBtGal35A

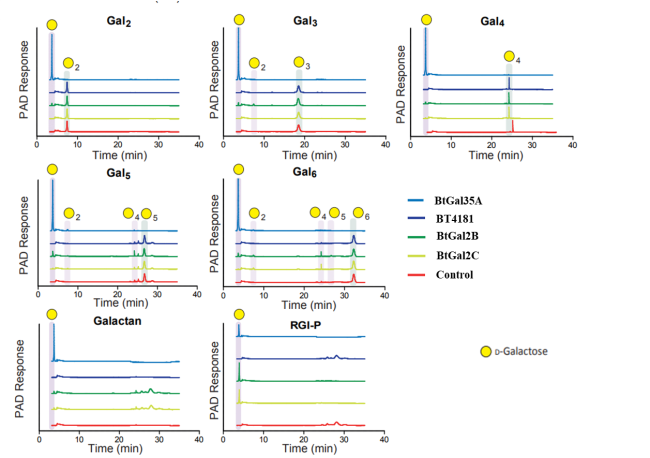
Figure 2. Analysis of the influence of galactosidase activity. The galactooligosaccharides at 1 mM, were incubated with 1 μM of enzyme for 16 h using standard conditions and the reactions were analysed by HPAEC. The data presented are representative of biological replicates n = 4.
4.PHYSICOCHEMICAL PROPERTIES
pH Optima: 7.5
Temperature Optima:37°C
5.STORAGE CONDITIONS
The enzyme should be stored at -20°C. For assay, this enzyme should be diluted in phosphate buffer (20 mM) pH 7.0. Swirl to mix the enzyme immediately prior to use.
6. REFERENCES
[1] Luis AS, Briggs J, Zhang X, et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol. 2018 Feb;3(2):210-219.


